Table of contents
- Introduction
- Packages
- Hyperparameter Tuning end-to-end process
- Reminder of Part 3 results
- Cross-validation plan
- Setup parallel processing
- Prophet Boost
- Prophet Boost summary results
- Random Forest summary results
- XGBoost summary results
- Prophet summary results
- Modeltime & Calibration tables
- Model evaluation results
- Forecast plot
- Save your work
- Conclusion
- References
Cover photo by christian buehner on Unsplash
Go to R-bloggers for R news and tutorials contributed by hundreds of R bloggers.
Introduction
This is the fourth of a series of 6 articles about time series forecasting with panel data and ensemble stacking with R.
Through these articles I will be putting into practice what I have learned from the Business Science University training course 1 "DS4B 203-R: High-Performance Time Series Forecasting", delivered by Matt Dancho. If you are looking to gain a high level of expertise in time series with R I strongly recommend this course.
The objective of this article is to explain the end-to-end process of time series hyperparamter tuning for non-sequential machine learning models like Random Forest, XGBoost, Prophet, and Prophet Boost. k-fold cross-validation can be applied to non-sequential algorithms.
You will understand how to tune parameters for Prophet Boost by performing several tuning rounds to adjust and stabilize the performance of the model. The hyperparameter tuning process is the same for the other algorithms.
The provided code is highly redundant for the sake of clarity. There is a lot of room for optimization with custom functions.
Prerequisites
I assume you are already familiar with the following topics, packages and terms:
dplyr or tidyverse R packages
preprocessing recipes from Part 2
model specification and workflows from Part 3
k-fold cross-validation
parameters of Random Forest, XGBoost, Prophet, and Prophet Boost algorithms
grid search
model evaluation metrics: RMSE, R-squared, ...
Packages
The following packages must be loaded:
# 01 FEATURE ENGINEERING
library(tidyverse) # loading dplyr, tibble, ggplot2, .. dependencies
library(timetk) # using timetk plotting, diagnostics and augment operations
library(tsibble) # for month to Date conversion
library(tsibbledata) # for aus_retail dataset
library(fastDummies) # for dummyfying categorical variables
library(skimr) # for quick statistics
# 02 FEATURE ENGINEERING WITH RECIPES
library(tidymodels) # with workflow dependency
# 03 MACHINE LEARNING
library(modeltime) # ML models specifications and engines
library(tictoc) # measure training elapsed time
# 04 HYPERPARAMETER TUNING
library(future)
library(doFuture)
library(plotly)
Hyperparameter Tuning end-to-end process
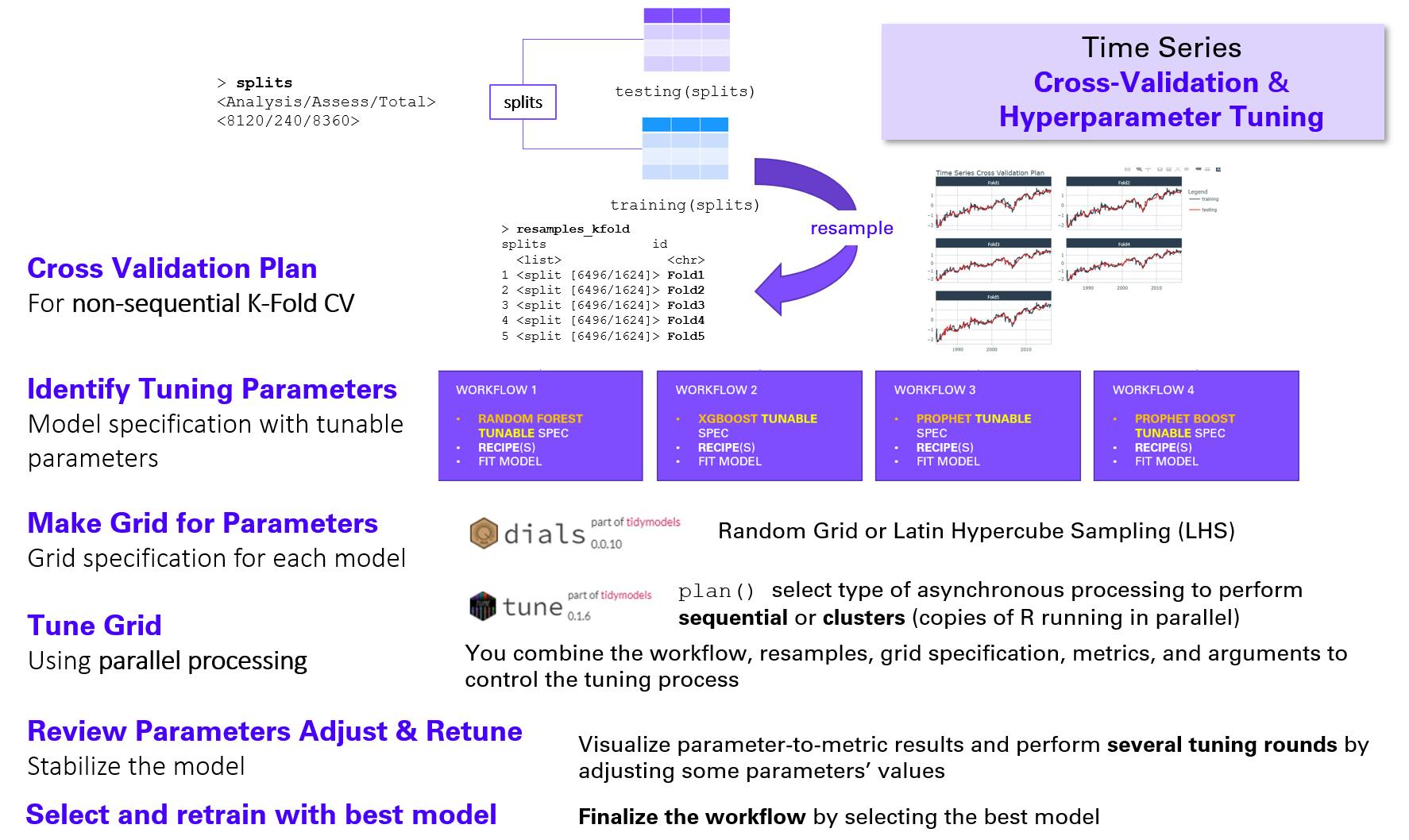
The end-to-end process is as follows:
Get the resamples. Here we will perform a k-fold cross-validation and obtain a cross-validation plan that we can plot to see "inside the folds".
Prepare for parallel process: register to future and get the number of vCores.
Define the tunable model specification: explicitly indicate the tunable parameters.
Define a tunable workflow into which we add the tunable model specification and the existing recipe; update the recipe if necessary.
Verify that the system has all tunable parameters' range, some parameter such as
mtrydo not have its values range correctly initialized.Define the search grid specification: you must provide type of grid (random or latin hypercube sampling) and its size.
Toggle on the parallel processing
Run the hyperparameter tuning by provding the tunable workflow, the resamples, and the grid specification.
Run several tuning rounds if you can e.g., if you have the time and resources and/or you are able to fix some parameter ranges.
After several tuning rounds, select the best model: with the lowest RMSE or with the higest R-squared, it may be same model or they can be different models.
(Re)fit the best model(s) with the training data by providing the tuned workflow.
Reminder of Part 3 results
Let us recall the non-tuned model evaluation results for the 4 machine learning algorithms:
# Load saved work from Part 3
> wflw_artifacts <- read_rds("workflows_artifacts_list.rds")
> wflw_artifacts$calibration$calibration_tbl %>%
modeltime_accuracy(testing(splits)) %>%
arrange(rmse)
# A tibble: 4 x 9
.model_id .model_desc .type mae mape mase smape rmse rsq
<int> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 3 PROPHET W/ REGRESSORS Test 0.213 22.9 0.470 22.9 0.267 0.846
2 4 PROPHET W/ XGBOOST ERRORS Test 0.203 27.1 0.448 20.6 0.283 0.750
3 2 XGBOOST Test 0.228 26.9 0.503 22.7 0.306 0.761
4 1 RANGER Test 0.244 25.6 0.538 25.3 0.322 0.763
Note: "RANGER" is the default engine for Random Forest modeltime::rand_forest().
Cross-validation plan
A k-fold cross-validation will randomly split the training data into k groups of roughly equal size (called "folds"). A resample of the analysis data consisted of k-1 of the folds while the assessment set contains the final fold. In basic k-fold cross-validation (i.e. no repeats), the number of resamples is equal to k.
It is important to understand that you may perform a k-fold cross-validation for all four machine learning models since they are non-sequential models. This is not the case for other models such as ARIMA, ETS (exponential smoothing) and NNETAR for which you should perform a time_series_cv() cross-validation.
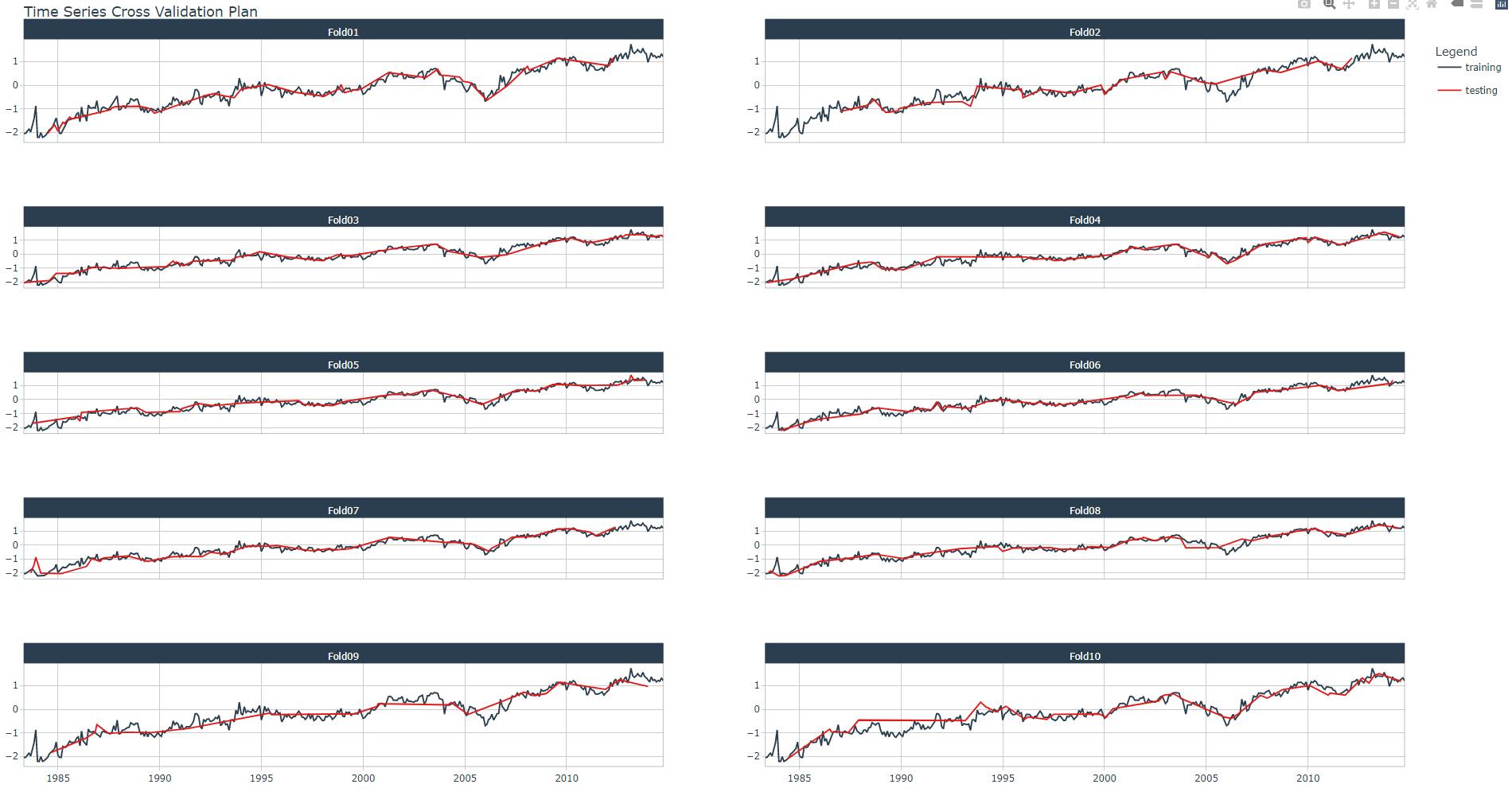
Perform the resample with function rsample::vfold_cv() as shown in the code snippet below. You may also display the cross validation plan and its plot (here, for one Industry) with tk_time_series_cv_plan() and plot_time_series_cv_plan(), respectively.
# k = 10 folds
> set.seed(123)
> resamples_kfold <- training(splits) %>%
vfold_cv(v = 10)
resamples_kfold %>%
tk_time_series_cv_plan() %>%
filter(Industry == Industries[1]) %>%
plot_time_series_cv_plan(.date_var = Month,
.value = Turnover,
.facet_ncol = 2)
Setup parallel processing
As per 1, "the future library allows for evaluating R expressions asynchronously. The doFuture package is an adapter that provides for running loops in parallel. The tune package manages the adapter process internally, so all we need to do is to set up doFuture".
# Registers the doFuture parallel processing
registerDoFuture()
# My laptop i5-8350U CPU has 8 threads (or vCores)
n_cores <- parallel::detectCores()
Prophet Boost
Prophet Boost - Identify tuning parameters
I've decided to tune the following parameters, you can run ?prophet_boost for a full description:
changepoint_num(prophet): Number of potential changepoints to include for modeling trend.mtry(xgboost): An integer for the number of predictors that will be randomly sampled at each split when creating the tree models.trees(xgboost): An integer for the number of trees contained in the ensemblemin_n(xgboost): An integer for the minimum number of data points in a node that are required for the node to be split further. tree_depth: An integer for the maximum depth of the tree (i.e. number of splits) (specific engines only).learn_rate(xgboost): A number for the rate at which the boosting algorithm adapts from iteration-to-iteration.loss_reduction(xgboost): A number for the reduction in the loss function required to split further.tree_depth(xgboost): An integer for the maximum depth of the tree (i.e. number of splits).
Define the tunable parameters with tune() within the model specification of prophet_boost(). Then, define a new tunable workflow for Prophet Boost (wflw_spec_rf_tune) by adding the tunable specification and the same recipe as in Part 3. Display the summary of the workflow as shown in the code snippet below.
> model_spec_prophet_boost_tune <- prophet_boost(
mode = "regression",
# growth = NULL,
changepoint_num = tune(),
#changepoint_range = NULL,
seasonality_yearly = FALSE,
seasonality_weekly = FALSE,
seasonality_daily = FALSE,
# season = NULL,
# prior_scale_changepoints = NULL,
# prior_scale_seasonality = NULL,
# prior_scale_holidays = NULL,
# logistic_cap = NULL,
# logistic_floor = NULL,
mtry = tune(),
trees = tune(),
min_n = tune(),
tree_depth = tune(),
learn_rate = tune(),
loss_reduction = tune(),
# sample_size = NULL,
# stop_iter = NULL
) %>%
set_engine("prophet_xgboost")
> wflw_spec_prophet_boost_tune <- workflow() %>%
add_model(model_spec_prophet_boost_tune) %>%
add_recipe(artifacts$recipes$recipe_spec)
> wflw_spec_prophet_boost_tune
== Workflow ===================================================================================================================================
Preprocessor: Recipe
Model: prophet_boost()
-- Preprocessor -------------------------------------------------------------------------------------------------------------------------------
5 Recipe Steps
* step_other()
* step_timeseries_signature()
* step_rm()
* step_dummy()
* step_normalize()
-- Model --------------------------------------------------------------------------------------------------------------------------------------
PROPHET Regression Model Specification (regression)
Main Arguments:
changepoint_num = tune()
seasonality_yearly = FALSE
seasonality_weekly = FALSE
seasonality_daily = FALSE
mtry = tune()
trees = tune()
min_n = tune()
tree_depth = tune()
learn_rate = tune()
loss_reduction = tune()
Computational engine: prophet_xgboost
Prophet Boost - Grid spec
Check parameters' range
The first thing to do is to display the parameters of the model with extract_parameter_set_dials(): you must check if there is any parameter with missing information about its values range.
As shown below, nparam[?] means that values range is missing for mtry parameter.
> extract_parameter_set_dials(model_spec_prophet_boost_tune)
Collection of 3 parameters for tuning
identifier type object
mtry mtry nparam[?]
trees trees nparam[+]
min_n min_n nparam[+]
Model parameters needing finalization:
# Randomly Selected Predictors ('mtry')
See `?dials::finalize` or `?dials::update.parameters` for more information.
Check the number of predictors looking at the recipe's summary.
> artifacts$recipes$recipe_spec %>%
update_role(Month, new_role = "indicator") %>%
prep() %>%
summary() %>%
group_by(role) %>%
summarise(n=n())
# A tibble: 3 x 2
role n
<chr> <int>
1 indicator 1
2 outcome 1
3 predictor 50
The new range update will be applied within the grid specifications in the next code snippet.
Grid search specification
Thanks to the dials package it is possible to define specifications for either Random Grid or Latin Hypercube Sampling (LHS).
We define a grid of size 20 e.g. 20 random values combinations of the 7 tunable parameters. Remember to set the seed to fix the grid since it uses a random process, you may have different values.
Since we have defined a 10-fold cross-validation there will be 20 predictions per fold and the mean of the performance metric (RMSE, R-squared, ...) will be calculated.
set.seed(123)
grid_spec_1 <- grid_latin_hypercube(
extract_parameter_set_dials(model_spec_prophet_boost_tune) %>%
update(mtry = mtry(range = c(1, 50))),
size = 20
)
> grid_spec_1
# A tibble: 20 x 7
changepoint_num mtry trees min_n tree_depth learn_rate loss_reduction
<int> <int> <int> <int> <int> <dbl> <dbl>
1 6 9 145 26 5 2.50e- 7 1.89e- 6
2 33 33 1538 18 14 5.55e- 6 2.17e- 2
3 8 42 42 21 3 2.22e- 6 1.63e- 1
4 0 23 1183 4 12 2.44e- 3 1.91e-10
5 30 30 1261 25 11 1.34e- 9 1.90e- 8
6 22 5 1882 3 7 1.45e- 8 1.01e- 7
7 45 48 971 30 6 2.94e-10 5.34e- 3
8 20 7 798 27 15 1.18e- 4 4.65e- 9
9 14 22 1993 12 3 6.54e- 7 1.42e- 4
10 27 37 1620 35 8 3.16e- 2 7.35e- 8
11 23 45 1450 8 4 6.11e- 4 7.60e- 2
12 31 1 1756 14 13 8.92e- 8 2.41e- 3
13 37 19 402 39 9 2.28e- 9 4.83e- 4
14 39 46 245 31 11 2.60e- 8 9.67e- 7
15 42 13 336 37 2 6.48e- 3 5.83e-10
16 17 28 1311 34 8 2.41e-10 2.92e- 5
17 49 31 870 11 2 2.32e- 5 1.70e+ 1
18 4 13 575 8 13 2.68e- 4 1.09e+ 0
19 44 18 1097 20 10 3.33e- 5 5.98e+ 0
20 11 40 613 16 6 5.62e- 2 7.32e- 6
Prophet Boost - Tune Grid
Let us perform 3 tuning rounds with 3 different grid specifications. I will explain at each round why do we perform the corresponding grid spec:
Round 1 with
grid_spec_1Round 2 with
grid_spec_2Round 3 with
grid_spec_3
To tune the workflow with a grid you need to provide the resamples, the grid specification and some controls on the grid search process.
tic()
tune_results_prophet_boost_1 <- wflw_spec_prophet_boost_tune %>%
tune_grid(
resamples = resamples_kfold,
grid = grid_spec_1,
control = control_grid(verbose = TRUE,
allow_par = TRUE)
)
toc()
> plan(strategy = sequential)
> tune_results_prophet_boost_1 %>%
+ show_best("rmse", n = Inf)
# A tibble: 20 x 13
changepoint_num mtry trees min_n tree_depth learn_rate loss_reduction .metric .estimator mean n std_err .config
<int> <int> <int> <int> <int> <dbl> <dbl> <chr> <chr> <dbl> <int> <dbl> <chr>
1 11 40 613 16 6 5.62e- 2 7.32e- 6 rmse standard 0.185 10 0.00372 Preprocessor~
2 27 37 1620 35 8 3.16e- 2 7.35e- 8 rmse standard 0.187 10 0.00378 Preprocessor~
3 0 23 1183 4 12 2.44e- 3 1.91e-10 rmse standard 0.192 10 0.00422 Preprocessor~
4 42 13 336 37 2 6.48e- 3 5.83e-10 rmse standard 0.308 10 0.00407 Preprocessor~
5 23 45 1450 8 4 6.11e- 4 7.60e- 2 rmse standard 0.360 10 0.00406 Preprocessor~
6 4 13 575 8 13 2.68e- 4 1.09e+ 0 rmse standard 0.550 10 0.00560 Preprocessor~
7 20 7 798 27 15 1.18e- 4 4.65e- 9 rmse standard 0.579 10 0.00588 Preprocessor~
8 44 18 1097 20 10 3.33e- 5 5.98e+ 0 rmse standard 0.606 10 0.00605 Preprocessor~
9 49 31 870 11 2 2.32e- 5 1.70e+ 1 rmse standard 0.614 10 0.00619 Preprocessor~
10 33 33 1538 18 14 5.55e- 6 2.17e- 2 rmse standard 0.619 10 0.00617 Preprocessor~
11 14 22 1993 12 3 6.54e- 7 1.42e- 4 rmse standard 0.623 10 0.00619 Preprocessor~
12 17 28 1311 34 8 2.41e-10 2.92e- 5 rmse standard 0.623 10 0.00620 Preprocessor~
13 39 46 245 31 11 2.60e- 8 9.67e- 7 rmse standard 0.623 10 0.00620 Preprocessor~
14 31 1 1756 14 13 8.92e- 8 2.41e- 3 rmse standard 0.623 10 0.00621 Preprocessor~
15 22 5 1882 3 7 1.45e- 8 1.01e- 7 rmse standard 0.623 10 0.00621 Preprocessor~
16 37 19 402 39 9 2.28e- 9 4.83e- 4 rmse standard 0.623 10 0.00620 Preprocessor~
17 8 42 42 21 3 2.22e- 6 1.63e- 1 rmse standard 0.623 10 0.00617 Preprocessor~
18 45 48 971 30 6 2.94e-10 5.34e- 3 rmse standard 0.623 10 0.00619 Preprocessor~
19 30 30 1261 25 11 1.34e- 9 1.90e- 8 rmse standard 0.623 10 0.00619 Preprocessor~
20 6 9 145 26 5 2.50e- 7 1.89e- 6 rmse standard 0.624 10 0.00616 Preprocessor~
> tune_results_prophet_boost_1 %>%
+ show_best("rsq", n = Inf)
# A tibble: 20 x 13
changepoint_num mtry trees min_n tree_depth learn_rate loss_reduction .metric .estimator mean n std_err .config
<int> <int> <int> <int> <int> <dbl> <dbl> <chr> <chr> <dbl> <int> <dbl> <chr>
1 11 40 613 16 6 5.62e- 2 7.32e- 6 rsq standard 0.958 10 0.00191 Preprocessor~
2 27 37 1620 35 8 3.16e- 2 7.35e- 8 rsq standard 0.957 10 0.00195 Preprocessor~
3 0 23 1183 4 12 2.44e- 3 1.91e-10 rsq standard 0.956 10 0.00218 Preprocessor~
4 23 45 1450 8 4 6.11e- 4 7.60e- 2 rsq standard 0.893 10 0.00325 Preprocessor~
5 42 13 336 37 2 6.48e- 3 5.83e-10 rsq standard 0.888 10 0.00293 Preprocessor~
6 4 13 575 8 13 2.68e- 4 1.09e+ 0 rsq standard 0.855 10 0.00460 Preprocessor~
7 20 7 798 27 15 1.18e- 4 4.65e- 9 rsq standard 0.845 10 0.00511 Preprocessor~
8 44 18 1097 20 10 3.33e- 5 5.98e+ 0 rsq standard 0.835 10 0.00535 Preprocessor~
9 33 33 1538 18 14 5.55e- 6 2.17e- 2 rsq standard 0.832 10 0.00551 Preprocessor~
10 49 31 870 11 2 2.32e- 5 1.70e+ 1 rsq standard 0.832 10 0.00549 Preprocessor~
11 14 22 1993 12 3 6.54e- 7 1.42e- 4 rsq standard 0.830 10 0.00555 Preprocessor~
12 17 28 1311 34 8 2.41e-10 2.92e- 5 rsq standard 0.830 10 0.00557 Preprocessor~
13 39 46 245 31 11 2.60e- 8 9.67e- 7 rsq standard 0.830 10 0.00557 Preprocessor~
14 22 5 1882 3 7 1.45e- 8 1.01e- 7 rsq standard 0.830 10 0.00557 Preprocessor~
15 45 48 971 30 6 2.94e-10 5.34e- 3 rsq standard 0.830 10 0.00556 Preprocessor~
16 37 19 402 39 9 2.28e- 9 4.83e- 4 rsq standard 0.830 10 0.00558 Preprocessor~
17 8 42 42 21 3 2.22e- 6 1.63e- 1 rsq standard 0.830 10 0.00554 Preprocessor~
18 30 30 1261 25 11 1.34e- 9 1.90e- 8 rsq standard 0.830 10 0.00557 Preprocessor~
19 31 1 1756 14 13 8.92e- 8 2.41e- 3 rsq standard 0.830 10 0.00557 Preprocessor~
20 6 9 145 26 5 2.50e- 7 1.89e- 6 rsq standard 0.829 10 0.00552 Preprocessor~
Round 1
Plot the tuning results for analysis. You may pipe the tuning results into feasts::autoplot() and generated and plot an interactive ggplot object with ggplotly, as shown in the code snippet below:
gr1<- tune_results_prophet_boost %>%
autoplot() +
geom_smooth(se = FALSE)
ggplotly(gr1)
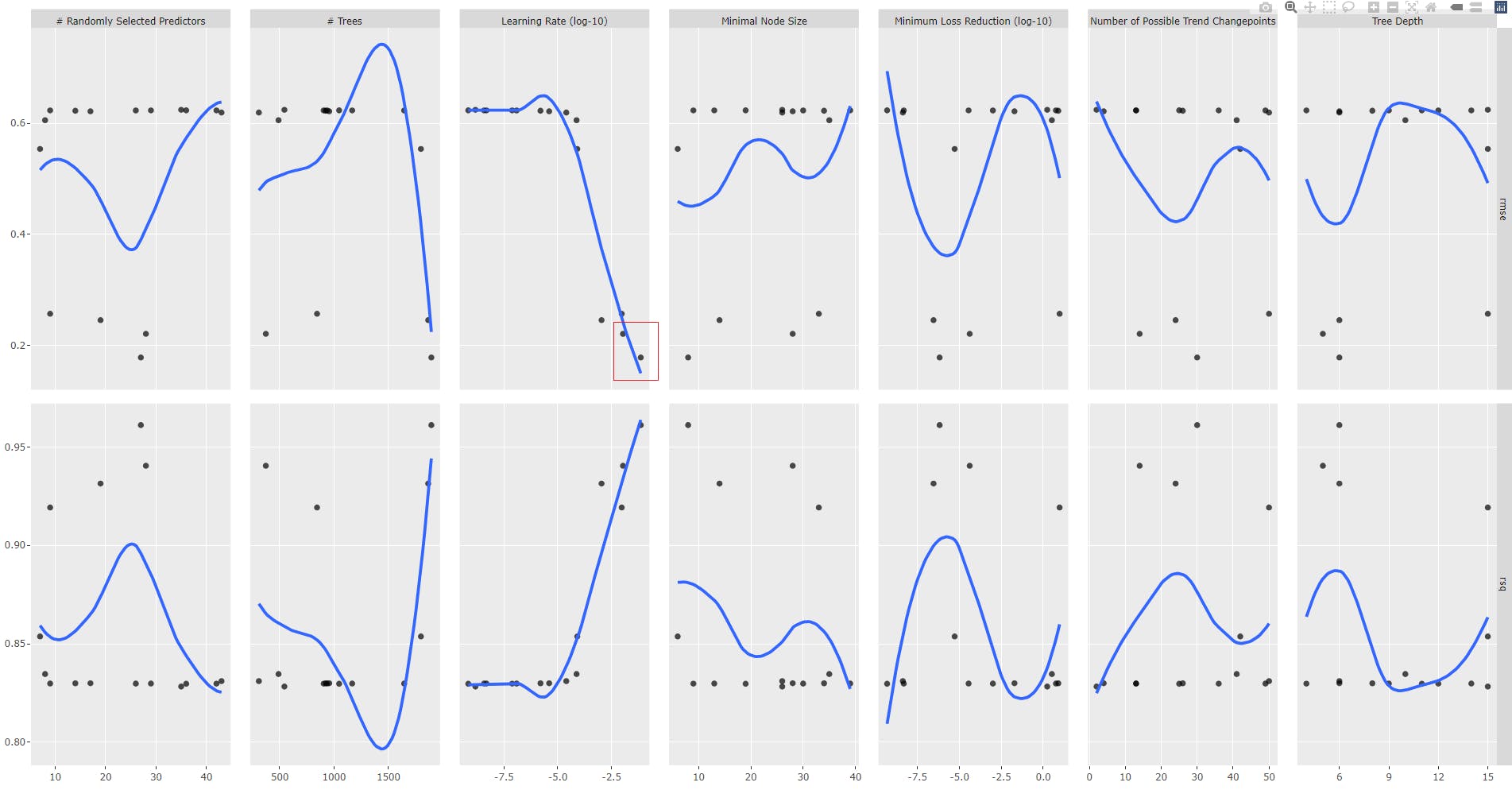
This is the most important part of the process: as a data scientist, you will decide of the tuning strategy.
Try to spot the plot with a monotonic smooth fonction (blue line), descending for RMSE or ascending for R-squared. Here, it corresponds to "Learning Rate (log-10)" parameter. Update the grid spec with a new range of values for Learning Rate where the RMSE is minimal.
Generally speaking we will do the following steps for each tuning round
update or adjust the parameter range within the grid specification.
toggle on parallel processing
perform hyperparameter tuning with new grid specification
toggle off parallel processing
analyze best RMSE and RSQ results
go to 1.
Round 2
We fix learn_rate parameter range of values.
# 1. update or adjust the parameter range within the grid specification.
set.seed(123)
grid_spec_2 <- grid_latin_hypercube(
extract_parameter_set_dials(model_spec_prophet_boost_tune) %>%
update(mtry = mtry(range = c(1, 50)),
learn_rate = learn_rate(range = c(-2.0, -1.0))),
size = 20
)
# 2. toggle on parallel processing
plan(
strategy = cluster,
workers = parallel::makeCluster(n_cores)
)
# 3. perform hyperparameter tuning with new grid specification
tic()
tune_results_prophet_boost_2 <- wflw_spec_prophet_boost_tune %>%
tune_grid(
resamples = resamples_kfold,
grid = grid_spec_2,
control = control_grid(verbose = TRUE,
allow_par = TRUE)
)
toc()
# 4. toggle off parallel processing
plan(strategy = sequential)
# 5. analyze best RMSE and RSQ results (here top 2)
> tune_results_prophet_boost_2 %>%
+ show_best("rsq", n = 2)
# A tibble: 2 x 13
changepoint_num mtry trees min_n tree_depth learn_rate loss_reduction .metric .estimator mean n std_err .config
<int> <int> <int> <int> <int> <dbl> <dbl> <chr> <chr> <dbl> <int> <dbl> <chr>
1 0 23 1183 4 12 0.0662 1.91e-10 rsq standard 0.961 10 0.00217 Preprocessor1~
2 11 40 613 16 6 0.0938 7.32e- 6 rsq standard 0.960 10 0.00171 Preprocessor1~
> tune_results_prophet_boost_2 %>%
+ show_best("rmse", n = 2)
# A tibble: 2 x 13
changepoint_num mtry trees min_n tree_depth learn_rate loss_reduction .metric .estimator mean n std_err .config
<int> <int> <int> <int> <int> <dbl> <dbl> <chr> <chr> <dbl> <int> <dbl> <chr>
1 0 23 1183 4 12 0.0662 1.91e-10 rmse standard 0.179 10 0.00462 Preprocessor1~
2 11 40 613 16 6 0.0938 7.32e- 6 rmse standard 0.181 10 0.00340 Preprocessor1~
Let us analyse the plot
gr2 <- tune_results_prophet_boost_2 %>%
autoplot() +
geom_smooth(se = FALSE)
ggplotly(gr2)
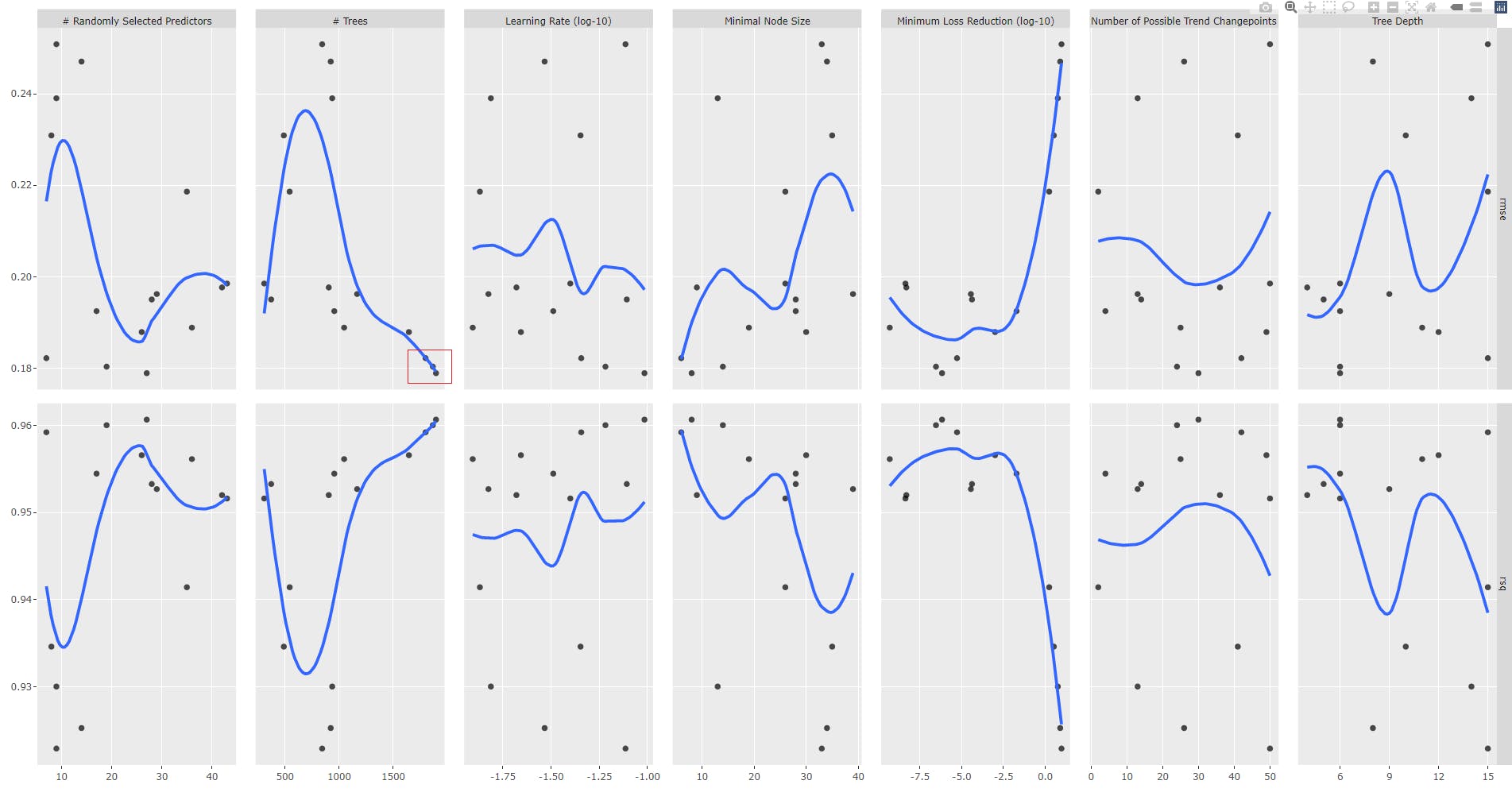
Round 3
We fix trees parameter range of values.
set.seed(123)
grid_spec_3 <- grid_latin_hypercube(
extract_parameter_set_dials(model_spec_prophet_boost_tune) %>%
update(mtry = mtry(range = c(1, 50)),
learn_rate = learn_rate(range = c(-2.0, -1.0)),
trees = trees(range = c(1183, 1993))),
size = 20
)
plan(
strategy = cluster,
workers = parallel::makeCluster(n_cores)
)
tic()
tune_results_prophet_boost_3 <- wflw_spec_prophet_boost_tune %>%
tune_grid(
resamples = resamples_kfold,
grid = grid_spec_3,
control = control_grid(verbose = TRUE,
allow_par = TRUE)
)
toc()
plan(strategy = sequential)
> tune_results_prophet_boost_3 %>%
+ show_best("rmse", n = 2)
# A tibble: 2 x 13
changepoint_num mtry trees min_n tree_depth learn_rate loss_reduction .metric .estimator mean n std_err .config
<int> <int> <int> <int> <int> <dbl> <dbl> <chr> <chr> <dbl> <int> <dbl> <chr>
1 0 23 1662 4 12 0.0662 1.91e-10 rmse standard 0.179 10 0.00464 Preprocessor1~
2 11 40 1431 16 6 0.0938 7.32e- 6 rmse standard 0.179 10 0.00363 Preprocessor1~
> tune_results_prophet_boost_3 %>%
+ show_best("rsq", n = 2)
# A tibble: 2 x 13
changepoint_num mtry trees min_n tree_depth learn_rate loss_reduction .metric .estimator mean n std_err .config
<int> <int> <int> <int> <int> <dbl> <dbl> <chr> <chr> <dbl> <int> <dbl> <chr>
1 11 40 1431 16 6 0.0938 7.32e- 6 rsq standard 0.961 10 0.00174 Preprocessor1~
2 0 23 1662 4 12 0.0662 1.91e-10 rsq standard 0.961 10 0.00220 Preprocessor1~
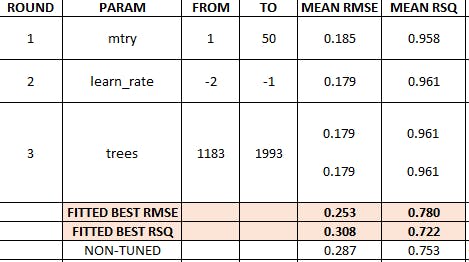
We obtain two best models, they show the same RMSE and RSQ values because there are rounded figures.
Select and fit the best model(s)
Please recall that we deal with workflows (not models directly) which incoporate a a model and a preprocessing recipe.
We finalize (finalize_workflow()) the tuning process by selecting the best model (select_best()) from round 3 tuning results (tune_results_prophet_boost_3) and finally, we refit the model (fit(training(splits))), and display accuracy results.
# Fitting round 3 best RMSE model
set.seed(123)
wflw_fit_prophet_boost_tuned <- wflw_spec_prophet_boost_tune %>%
finalize_workflow(
select_best(tune_results_prophet_boost_3, "rmse", n=1)) %>%
fit(training(splits))
modeltime_table(wflw_fit_prophet_boost_tuned) %>%
modeltime_calibrate(testing(splits)) %>%
modeltime_accuracy()
# A tibble: 1 x 9
.model_id .model_desc .type mae mape mase smape rmse rsq
<int> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 1 PROPHET W/ XGBOOST ERRORS Test 0.191 25.7 0.421 19.9 0.253 0.780
# Fitting round 3 best RSQmodel
set.seed(123)
wflw_fit_prophet_boost_tuned_rsq <- wflw_spec_prophet_boost_tune %>%
finalize_workflow(
select_best(tune_results_prophet_boost_3, "rsq", n=1)) %>%
fit(training(splits))
modeltime_table(wflw_fit_prophet_boost_tuned_rsq) %>%
modeltime_calibrate(testing(splits)) %>%
modeltime_accuracy()
# A tibble: 1 x 9
.model_id .model_desc .type mae mape mase smape rmse rsq
<int> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 1 PROPHET W/ XGBOOST ERRORS Test 0.223 26.2 0.492 22.3 0.308 0.722
As shown above, the best fitted model is the first one, thus we select and save it.
Save Prophet Boot tuning artifacts
It is very helpful the save all artifacts of the tuning process. I've created a saved to an RDS file a R list for which the structure should the same for all algorithms.
tuned_prophet_xgb <- list(
# Workflow spec
tune_wkflw_spec = wflw_spec_prophet_boost_tune,
# Grid spec
tune_grid_spec = list(
round1 = grid_spec_1,
round2 = grid_spec_2,
round3 = grid_spec_3),
# Tuning Results
tune_results = list(
round1 = tune_results_prophet_boost_1,
round2 = tune_results_prophet_boost_2,
round3 = tune_results_prophet_boost_3),
# Tuned Workflow Fit
tune_wflw_fit = wflw_fit_prophet_boost_tuned,
# from FE
splits = artifacts$splits,
data = artifacts$data,
recipes = artifacts$recipes,
standardize = artifacts$standardize,
normalize = artifacts$normalize
)
tuned_prophet_xgb %>%
write_rds("tuning/tuned_prophet_xgb.rds")
Prophet Boost summary results
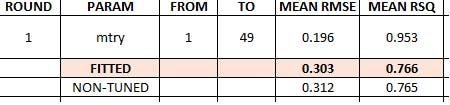
Random Forest summary results
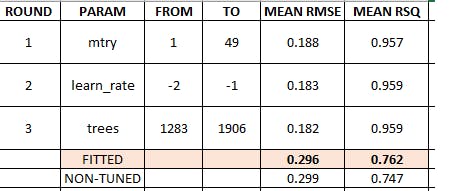
XGBoost summary results
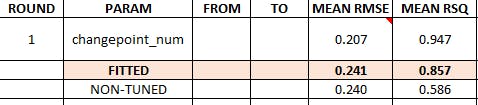
Prophet summary results
Modeltime & Calibration tables
The objective is to add all non-tuned and tuned models into a single modeltime table submodels_all_tbl.
Please recall that we deal with workflows (not models directly) which incoporates a model and a preprocessing recipe.
Notice how we combined (combine_modeltime_tables()) the tables and updated the description (update_model_description()) for the tuned workflows.
> submodels_tbl <- modeltime_table(
wflw_artifacts$workflows$wflw_random_forest,
wflw_artifacts$workflows$wflw_xgboost,
wflw_artifacts$workflows$wflw_prophet,
wflw_artifacts$workflows$wflw_prophet_boost
)
> submodels_all_tbl <- modeltime_table(
tuned_rf_rds$tune_wflw_fit,
tuned_xgboost_rds$tune_wflw_fit,
tuned_prophet_rds$tune_wflw_fit,
tuned_prophet_fou_boost_rds$tune_wflw_fit
) %>%
update_model_description(1, "RANGER - Tuned") %>%
update_model_description(2, "XGBOOST - Tuned") %>%
update_model_description(3, "PROPHET W/ REGRESSORS - Tuned") %>%
update_model_description(4, "PROPHET W/ XGBOOST ERRORS - Tuned") %>%
combine_modeltime_tables(submodels_tbl)
> submodels_all_tbl
# Modeltime Table
# A tibble: 8 x 3
.model_id .model .model_desc
<int> <list> <chr>
1 1 <workflow> RANGER - Tuned
2 2 <workflow> XGBOOST - Tuned
3 3 <workflow> PROPHET W/ REGRESSORS - Tuned
4 4 <workflow> PROPHET W/ XGBOOST ERRORS - Tuned
5 5 <workflow> RANGER
6 6 <workflow> XGBOOST
7 7 <workflow> PROPHET W/ REGRESSORS
8 8 <workflow> PROPHET W/ XGBOOST ERRORS
Model evaluation results
We calibrate all fitted workflows with the test dataset and display the accuracy results for all non-tuned and tuned models. You may also create an interactive table which facilitates sorting, as shown in the code snippet below.
> calibration_all_tbl <- submodels_all_tbl %>%
modeltime_calibrate(testing(splits))
calibration_all_tbl %>%
modeltime_accuracy() %>%
arrange(rmse)
# A tibble: 8 x 9
.model_id .model_desc .type mae mape mase smape rmse rsq
<int> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 7 PROPHET W/ REGRESSORS Test 0.189 21.6 0.417 21.5 0.240 0.856
2 3 PROPHET W/ REGRESSORS - Tuned Test 0.189 21.6 0.418 21.5 0.241 0.857
3 4 PROPHET W/ XGBOOST ERRORS - Tuned Test 0.191 25.7 0.421 19.9 0.253 0.780
4 8 PROPHET W/ XGBOOST ERRORS Test 0.204 24.8 0.451 20.8 0.287 0.753
5 2 XGBOOST - Tuned Test 0.219 23.6 0.482 22.9 0.296 0.762
6 6 XGBOOST Test 0.216 25.0 0.476 22.4 0.299 0.747
7 1 RANGER - Tuned Test 0.231 26.1 0.509 24.6 0.303 0.766
8 5 RANGER Test 0.235 25.4 0.519 25.0 0.312 0.765
calibration_all_tbl %>%
modeltime_accuracy() %>%
arrange(desc(rsq))
# A tibble: 8 x 9
.model_id .model_desc .type mae mape mase smape rmse rsq
<int> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 3 PROPHET W/ REGRESSORS - Tuned Test 0.189 21.6 0.418 21.5 0.241 0.857
2 7 PROPHET W/ REGRESSORS Test 0.189 21.6 0.417 21.5 0.240 0.856
3 4 PROPHET W/ XGBOOST ERRORS - Tuned Test 0.191 25.7 0.421 19.9 0.253 0.780
4 1 RANGER - Tuned Test 0.231 26.1 0.509 24.6 0.303 0.766
5 5 RANGER Test 0.235 25.4 0.519 25.0 0.312 0.765
6 2 XGBOOST - Tuned Test 0.219 23.6 0.482 22.9 0.296 0.762
7 8 PROPHET W/ XGBOOST ERRORS Test 0.204 24.8 0.451 20.8 0.287 0.753
8 6 XGBOOST Test 0.216 25.0 0.476 22.4 0.299 0.747
# Interactive table (bonus !)
calibration_all_tbl %>%
modeltime_accuracy() %>%
table_modeltime_accuracy()
Best fitted models are Prophet (best RMSE) and tuned Prophet (best RSQ).
Forecast plot
We plot a forecast with test data using all models and industries, the figure below is a zoom of the test data predictions vs. actual data.
calibration_all_tbl %>%
modeltime_forecast(
new_data = testing(splits),
actual_data = artifacts$data$data_prepared_tbl,
keep_data = TRUE
) %>%
filter(Industry == Industries[1]) %>%
plot_modeltime_forecast(
#.facet_ncol = 4,
.conf_interval_show = FALSE,
.interactive = TRUE,
.title = Industries[1]
)
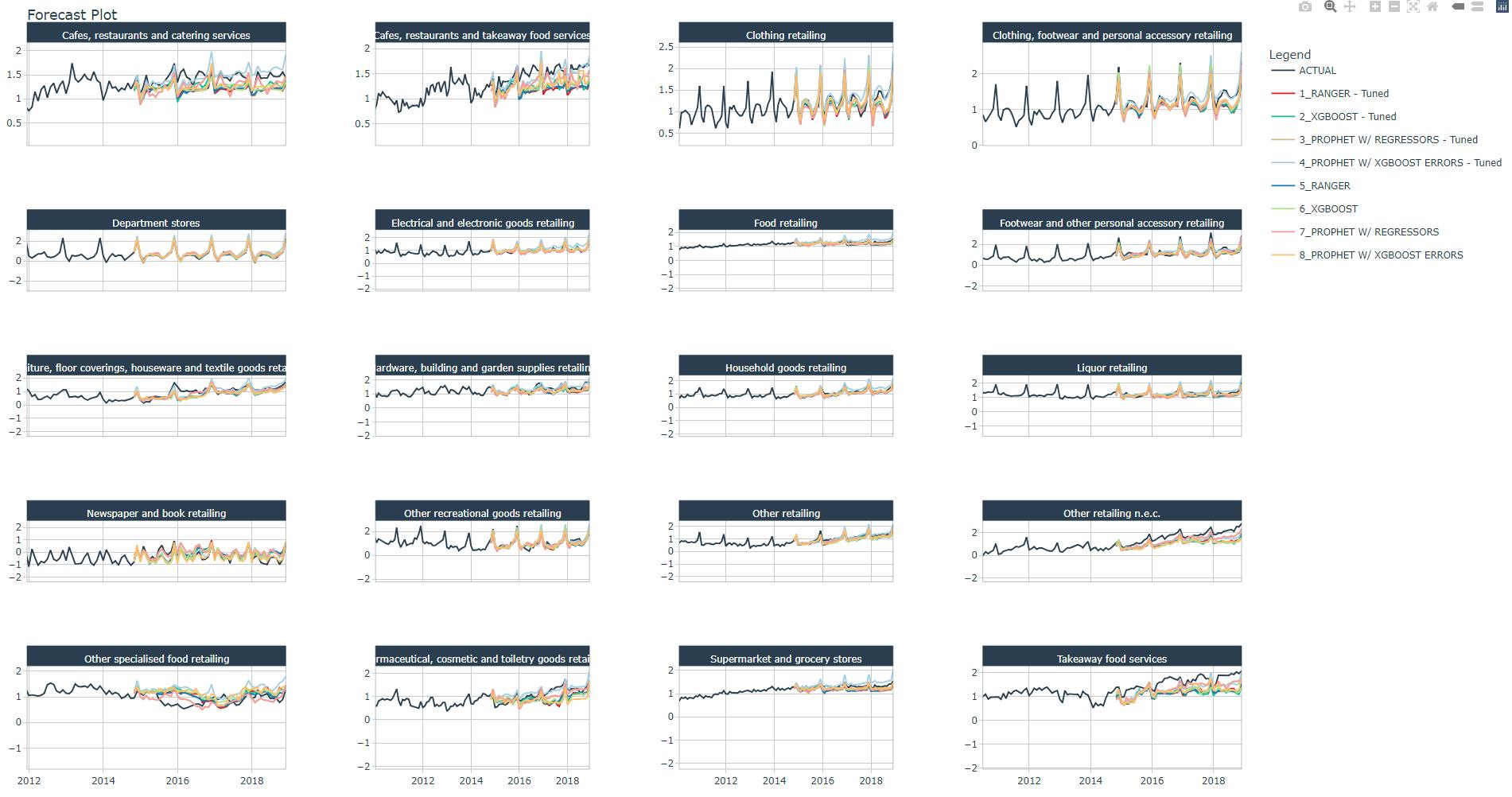
You may check and confirm the models that best follow the trend and spikes.
Save your work
workflow_all_artifacts <- list(
workflows = submodels_all_tbl,
calibration = calibration_all_tbl
)
workflow_all_artifacts %>%
write_rds("workflows_NonandTuned_artifacts_list.rds")
Conclusion
In this article you have learned how to perform hyperparameter tuning for 4 machine learning (non-sequentual) models: Random Forest, XGBoost, Prophet and Prohet Boost. A step-by-step detailed process was provided for Prophet Boost.
You have learned
how to define tunable specifications for each machine learning algorithm.
how to set up parallel processing with a cluster of vCores.
how to verify parameter values range prior tuning.
how to define a grid search specification for each algorithm
how to perform hyperparameter tuning and anlyse results with the plot against RMSE.
how to adjust a specific parameter and retune, performing several hyperparameter tuning rounds.
how to select the model and retrain the model.
how to add all tuned and non-tuned models into a modeltime and calibration table.
how to display all models accuracy results and plot forecast against the test dataset.
References
1 Dancho, Matt, "DS4B 203-R: High-Performance Time Series Forecasting", Business Science University

